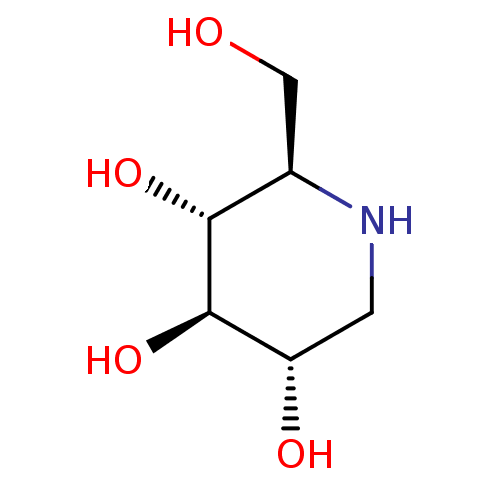
BDBM18351 (2R,3R,4R,5S)-2-(Hydroxymethyl)piperidine-3,4,5-triol, 10::(2R,3R,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol::1-Deoxynojirimycin::1-deoxynojirimycin (DNJ)::CHEMBL307429::US20230339856, Compound DNJ::US9181184, 1::dNM
SMILES OC[C@H]1NC[C@H](O)[C@@H](O)[C@@H]1O
InChI Key InChIKey=LXBIFEVIBLOUGU-JGWLITMVSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 18351
Found 3 hits for monomerid = 18351
Affinity DataIC50: 1.10E+5nMAssay Description:Inhibition of yeast maltase alpha-glucosidase assessed as p-nitrophenol release by spectrophotometricallyMore data for this Ligand-Target Pair
Affinity DataIC50: 4.26E+8nMAssay Description:Inhibition of yeast alpha-glucosidase using para-nitrophenyl alpha-D-glucopyranoside as substrate by spectrophotometric analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 2.78E+5nMAssay Description:Inhibition of yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate preincubated for 10 min before substrate addition and ...More data for this Ligand-Target Pair